An Overview of Image Registration
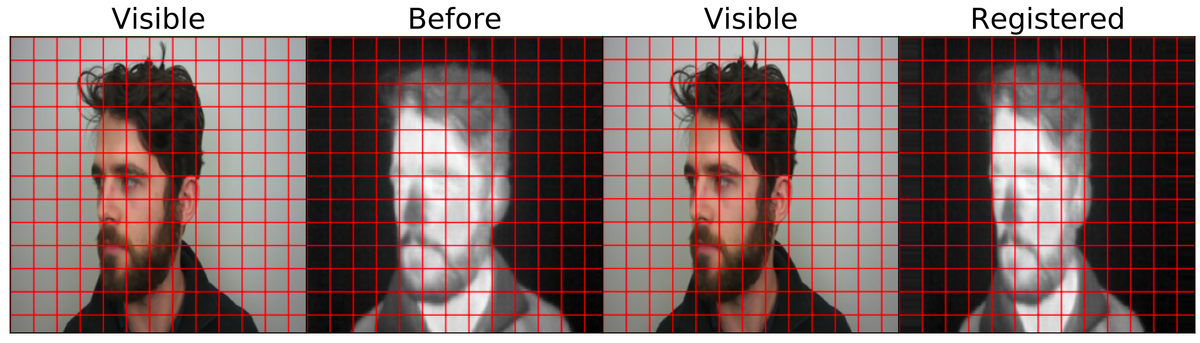
Image registration is a standard technique in computer vision, defined in the following ways: “directly overlaying one image on top of another” (Reddy et al), “aligning two or more images” (De Vos et al), and the “process of transforming different image datasets into one coordinate system with matched imaging contents” (Haskins et al).
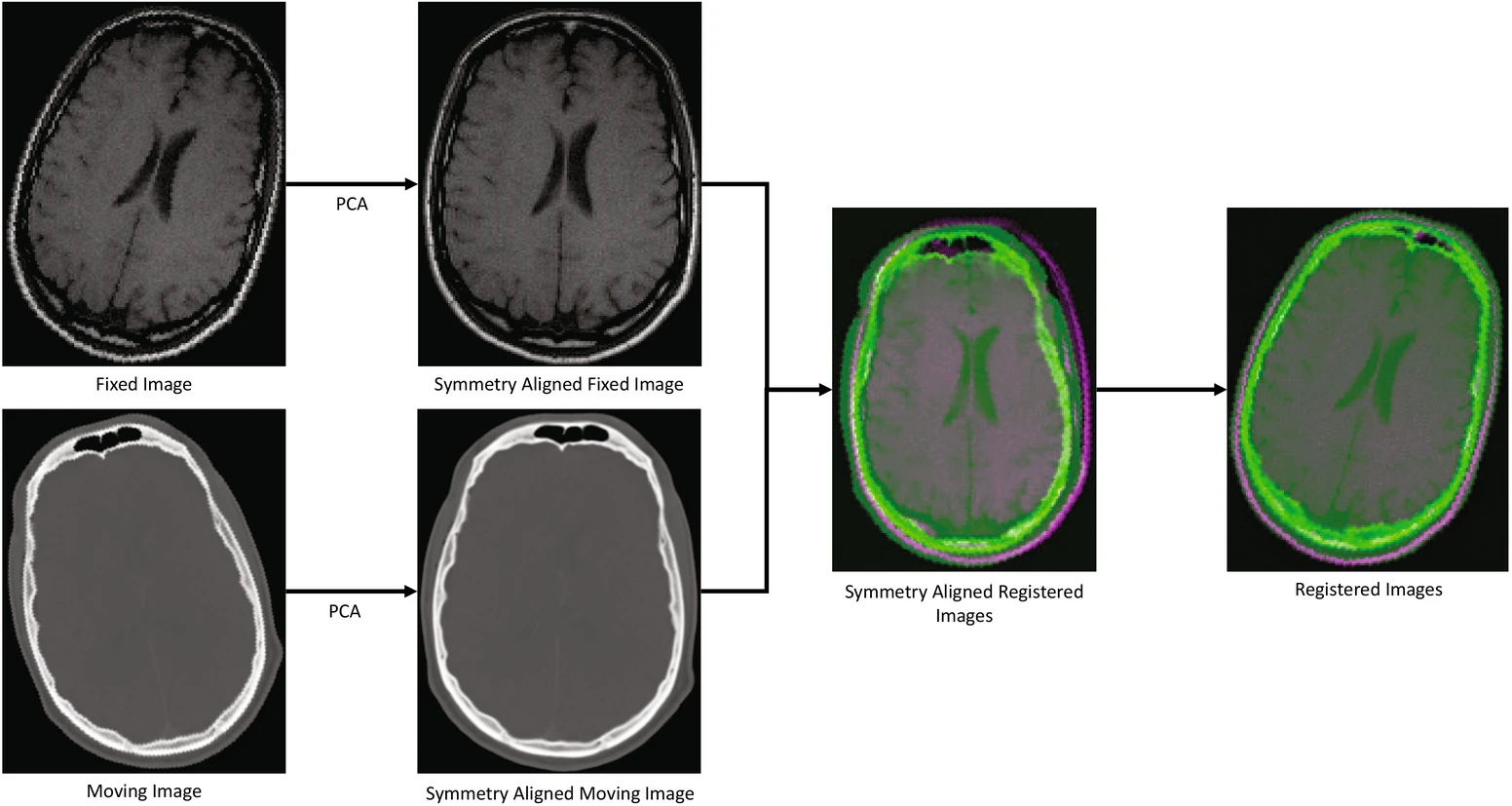
This is highly relevant in the biomedical domain where a “moving” image is aligned to match the scale of a “fixed” image. There are four steps in image registration, starting with feature detection, followed by feature matching, transform model estimation, and image resampling to align the image with its reference (Rao et al).
As such, image registration is used when one image, with respect to the other, is rotated, translated, or scaled differently.
On top of rotational problems, biomedical images are compounded by the fact that they are typically multi-modal (not operating only in the visible spectra), contain diverse and varied specimens, and exhibit non-distinct structures like microscopy (Lu, et al; Schnabel et al).
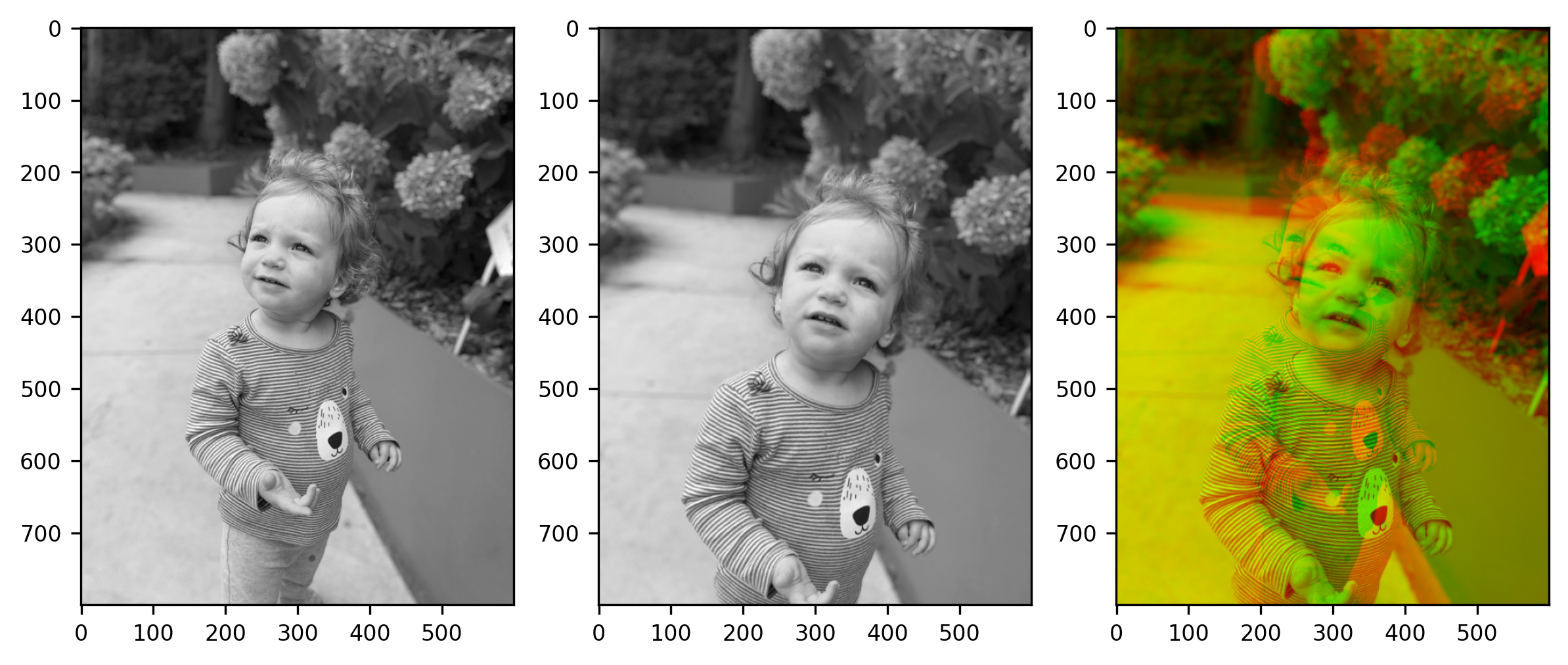
Reddy et al., describes three classes of conventional image registration models which are Fast Fourier Transform (FFT) algorithms, feature-based detectors like SIFT, or geometric models that steer rotation according to keypoints like ORB (Reddy et al). Recently, deep learning methods have been investigated to automatically register images by learning transformation parameters across images. For example, de Vos, et al., developed the Unsupervised Deep Learning Image Registration (DLIR), where registration can be performed in one pass using a multi-stage CNN that performs affine registration and deformable image registration in multiple phases (De Vos et al). DLIR predicts the transformation parameters which are used to calculate a dense displacement vector field. The field resamples the “moving” image into a similar “fixed” image matrix using Normalized Cross Correlation (NCC) as the similarity metric.
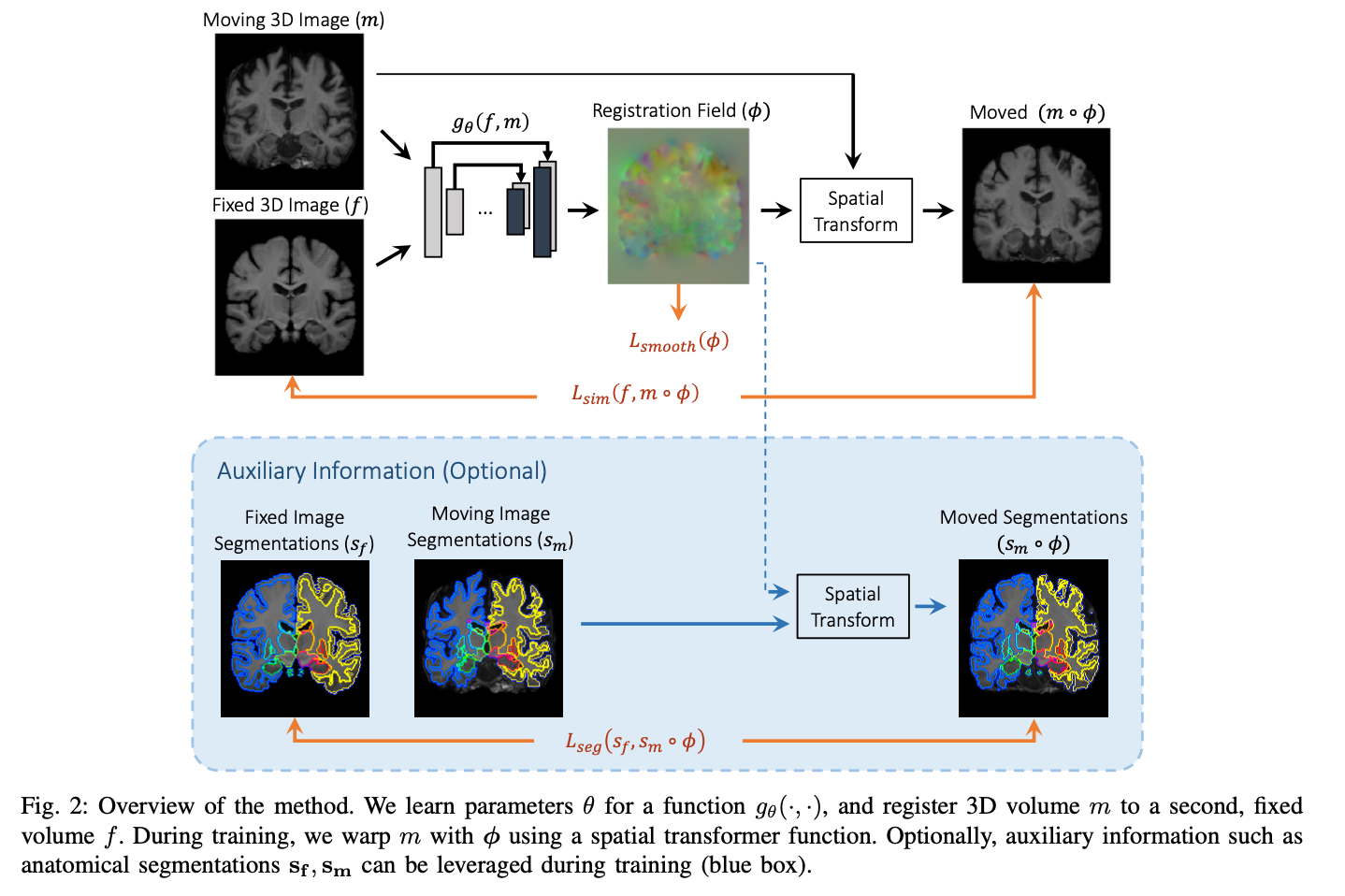
Many deep learning approaches like Voxelmorph (Balakrishnan et al), integrate a spatial transformation network (STN) (Jaderberg et al). The goal of the STN is to predict transformation parameters by a localization network to create a sampling grid. This grid consists of coordinates used to guide how the input feature map is sampled. An affine transformation can then be applied to the sampled vector to generate the registered output. The localization network can be any neural network like a CNN. A limited number of works explore the concurrent application of image registration with image-to-image translation.
In a recent survey by Lu et al., the authors investigate the success of applying image-to-image translation towards the biomedical image registration task. Xu et al., translates moving CT scans to fixed MRI scans, and registers both images by learning their deformation fields, with the goal of fusing them into a fixed MRI image (Xu, et al). In this work, both the ground truth CT and MRI scans are available to learn at the time of prediction.
Want to cite this? You can!
@book{ordun2023multimodal,
title={Multimodal Deep Generative Models for Cross-Spectral Image Analysis},
author={Ordun, Catherine Y},
year={2023},
publisher={University of Maryland, Baltimore County}
}
References:
B Srinivasa Reddy and Biswanath N Chatterji. An fft-based technique for transla- tion, rotation, and scale-invariant image registration. IEEE transactions on image processing, 5(8):1266–1271, 1996.
Bob D De Vos, Floris F Berendsen, Max A Viergever, Hessam Sokooti, Marius Staring, and Ivana Isˇgum. A deep learning framework for unsupervised affine and deformable image registration. Medical image analysis, 52:128–143, 2019.
Grant Haskins, Uwe Kruger, and Pingkun Yan. Deep learning in medical image registration: a survey. Machine Vision and Applications, 31(1):1–18, 2020.
Y Raghavender Rao, Nikhil Prathapani, and E Nagabhooshanam. Application of normalized cross correlation to image registration. International Journal of Research in Engineering and Technology, 3(5):12–16, 2014.
Jiahao Lu, et al. Is image-to- image translation the panacea for multimodal image registration? a comparative study. arXiv preprint arXiv:2103.16262, 2021.
Julia A Schnabel, Daniel Rueckert, Marcel Quist, Jane M Blackall, Andy D Castellano-Smith, Thomas Hartkens, Graeme P Penney, Walter A Hall, Haiying Liu, Charles L Truwit, et al. A generic framework for non-rigid registration based on non-uniform multi-level free-form deformations. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 573–581. Springer, 2001.
Guha Balakrishnan, Amy, Zhao,Mert R Sabuncu,John Guttag, and Adrian V Dalca. Voxelmorph: a learning framework for deformable medical image registration. IEEE transactions on medical imaging, 38(8):1788–1800, 2019.
Max Jaderberg, et al. Spatial transformer networks. NeurIPS, 28, 2015.
Zhe Xu et al. Adversarial uni-and multi-modal stream networks for multimodal image registration. In MICCAI, pages 222–232, 2020.